
Summary
The alpha and beta (or A and B) subunits are found in the F1, V1, and A1 complexes of F-, V- and A-ATPases, respectively, as well as flagellar (T3SS) ATPase and the termination factor Rho. The subunits make up a ring that contains the ATP-hydrolyzing (or producing) catalytic core. The F-ATPases (or F1Fo ATPases), V-ATPases (or V1Vo ATPases) and A-ATPases (or A1Ao ATPases) are composed of two linked complexes: the F1, V1 or A1 complex containsthat synthesizes/hydrolyses ATP, and the Fo, Vo or Ao complex that forms the membrane-spanning pore. The F-, V- and A-ATPases all contain rotary motors, one that drives proton translocation across the membrane and one that drives ATP synthesis/hydrolysis.[1][2]
| ATP synthase alpha/beta family, beta-barrel domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
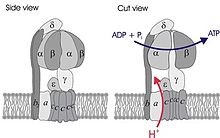 Simplified model of FOF1-ATPase alias ATP synthase of E. coli. Subunits of the enzyme are labeled accordingly. | |||||||||||
| Identifiers | |||||||||||
| Symbol | ATP-synt_ab_N | ||||||||||
| Pfam | PF02874 | ||||||||||
| InterPro | IPR004100 | ||||||||||
| PROSITE | PDOC00137 | ||||||||||
| SCOP2 | 1bmf / SCOPe / SUPFAM | ||||||||||
| |||||||||||
| ATP synthase alpha/beta family, nucleotide-binding domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | ATP-synt_ab | ||||||||||
| Pfam | PF00006 | ||||||||||
| InterPro | IPR000194 | ||||||||||
| PROSITE | PDOC00137 | ||||||||||
| SCOP2 | 1bmf / SCOPe / SUPFAM | ||||||||||
| |||||||||||
| ATP synthase alpha/beta chain, C terminal domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | ATP-synt_ab_C | ||||||||||
| Pfam | PF00306 | ||||||||||
| InterPro | IPR000793 | ||||||||||
| SCOP2 | 1bmf / SCOPe / SUPFAM | ||||||||||
| |||||||||||

ATPases (or ATP synthases) are membrane-bound enzyme complexes/ion transporters that combine ATP synthesis and/or hydrolysis with the transport of protons across a membrane. ATPases can harness the energy from a proton gradient, using the flux of ions across the membrane via the ATPase proton channel to drive the synthesis of ATP. Some ATPases work in reverse, using the energy from the hydrolysis of ATP to create a proton gradient.
There are different types of ATPases, which can differ in function (ATP synthesis and/or hydrolysis), structure (F-, V- and A-ATPases contain rotary motors) and in the type of ions they transport.[3][4] The types with this domain include:
- F-ATPases (F1Fo ATPases) are found in mitochondria, chloroplasts and bacterial plasma membranes are the prime producers of ATP, using the proton gradient generated by oxidative phosphorylation (mitochondria) or photosynthesis (chloroplasts).
- V-ATPases (V1Vo ATPases) are primarily found in eukaryotic vacuoles, catalysing ATP hydrolysis to transport solutes and lower pH in organelles.
- A-ATPases (A1Ao ATPases) are found in Archaea and function like F-ATPases.
- T3SS / flagellum ATPases, which are homologous to both parts of the A/F/V rotary ATPases: strongly in the "1" part, and weakly in the "O" part.[5]
- Ring-shaped DNA helicases like the Rho factor, where the ring is homologus to the α/β subunits.[6]
In F-ATPases, there are three copies each of the alpha and beta subunits that form the catalytic core of the F1 complex, while the remaining F1 subunits (gamma, delta, epsilon) form part of the stalks. There is a substrate-binding site on each of the alpha and beta subunits, those on the beta subunits being catalytic, while those on the alpha subunits are regulatory. The alpha and beta subunits form a cylinder that is attached to the central stalk. The alpha/beta subunits undergo a sequence of conformational changes leading to the formation of ATP from ADP, which are induced by the rotation of the gamma subunit, itself is driven by the movement of protons through the Fo complex C subunit.[7]
In V- and A-ATPases, the alpha/A and beta/B subunits of the V1 or A1 complex are homologous to the alpha and beta subunits in the F1 complex of F-ATPases, except that the alpha subunit is catalytic and the beta subunit is regulatory.
The alpha/A and beta/B subunits can each be divided into three regions, or domains, centred on the ATP-binding pocket, and based on structure and function. The central domain contains the nucleotide-binding residues that make direct contact with the ADP/ATP molecule.[8]
Human proteins containing this domain edit
References edit
- ^ Itoh H, Yoshida M, Yasuda R, Noji H, Kinosita K (2001). "Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase". Nature. 410 (6831): 898–904. Bibcode:2001Natur.410..898Y. doi:10.1038/35073513. PMID 11309608. S2CID 3274681.
- ^ Wilkens S, Zheng Y, Zhang Z (2005). "A structural model of the vacuolar ATPase from transmission electron microscopy". Micron. 36 (2): 109–126. doi:10.1016/j.micron.2004.10.002. PMID 15629643.
- ^ Muller V, Cross RL (2004). "The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio". FEBS Lett. 576 (1): 1–4. doi:10.1016/j.febslet.2004.08.065. PMID 15473999. S2CID 25800744.
- ^ Zhang X, Niwa H, Rappas M (2004). "Mechanisms of ATPases--a multi-disciplinary approach". Curr Protein Pept Sci. 5 (2): 89–105. doi:10.2174/1389203043486874. PMID 15078220.
- ^ Imada, Katsumi; Minamino, Tohru; Uchida, Yumiko; Kinoshita, Miki; Namba, Keiichi (29 March 2016). "Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator". Proceedings of the National Academy of Sciences. 113 (13): 3633–3638. Bibcode:2016PNAS..113.3633I. doi:10.1073/pnas.1524025113. PMC 4822572. PMID 26984495.
- ^ Skordalakes, Emmanuel; Berger, James M (July 2003). "Structure of the Rho Transcription Terminator". Cell. 114 (1): 135–146. doi:10.1016/S0092-8674(03)00512-9. PMID 12859904. S2CID 5765103.
- ^ Amzel LM, Bianchet MA, Leyva JA (2003). "Understanding ATP synthesis: structure and mechanism of the F1-ATPase (Review)". Mol. Membr. Biol. 20 (1): 27–33. doi:10.1080/0968768031000066532. PMID 12745923. S2CID 218895820.
- ^ Chandler D, Wang H, Antes I, Oster G (2003). "The unbinding of ATP from F1-ATPase". Biophys. J. 85 (2): 695–706. Bibcode:2003BpJ....85..695A. doi:10.1016/S0006-3495(03)74513-5. PMC 1303195. PMID 12885621.


